User Inputs
The simplest input to CoPilot is the ligand smiles along with a user-specific prompt (under the fields “Enter a SMILES String”, “Enter the desired Modifications” in Figure 1, respectively), this is particularly aimed at property optimisation and generates new molecules with improved target properties. The user provides a single input SMILES string. Pasting a correct SMILES string should produce a 2D image of the molecule, on the other hand, an incorrect SMILES will generate an error “Invalid SMILES string. Please enter a valid SMILES.” Along with the SMILES, the user is required to provide a prompt describing the desired modification for the molecule. These modifications can be broad, such as improving QED, or highly specific, such as modifying a specific chemical group on the molecule. Upon clicking “Generate Molecules”, CoPilot provides the user with five molecules with the desired modifications as requested by the user.
The user has the option to provide the protein-ligand complex as input, which provides on-the-fly docking scores for the generated ligands. The user is required to input the .pdb file of the protein-ligand complex, the residue name of the ligand as given in the PDB, and the ID of the protein chain to which the ligand is bound (under the fields “Protein File”, “Residue Name” and “Protein Chain” on Figure 1, respectively). This, along with the SMILES string allows the user to carry out ligand modifications and improve lead-likeliness while keeping in check the binding affinity by means of the docking score.
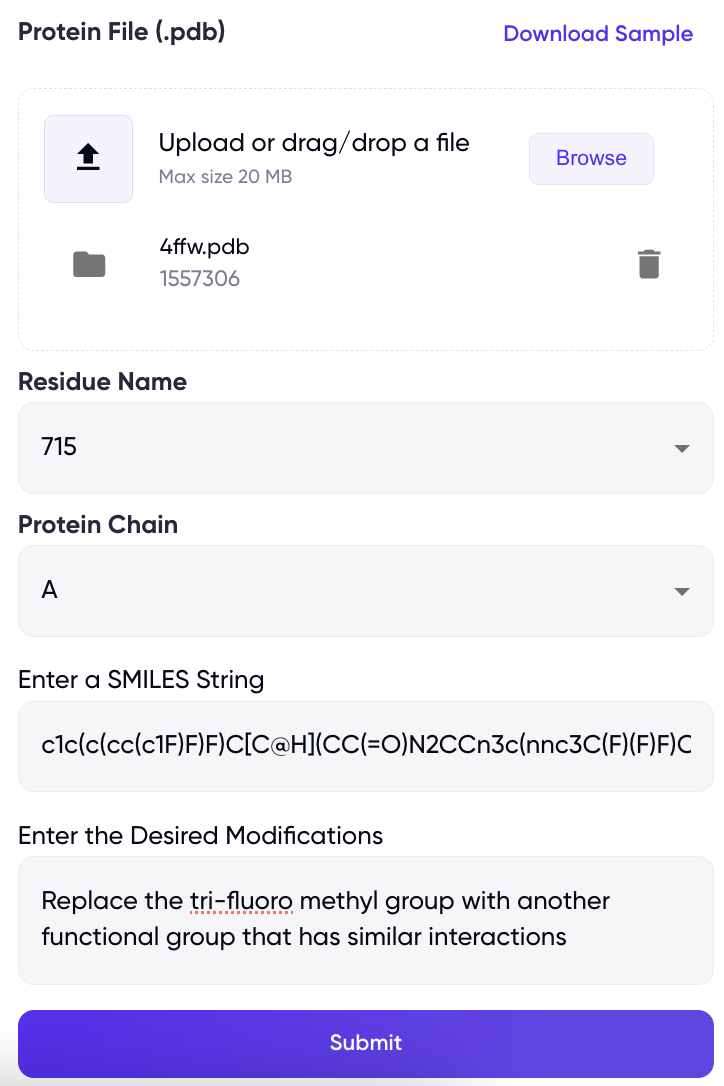
Figure 1. Primary user interface for CoPilot lead optimizer displaying the input fields.