User Inputs
The user inputs required for a MAIMol job are shown below.
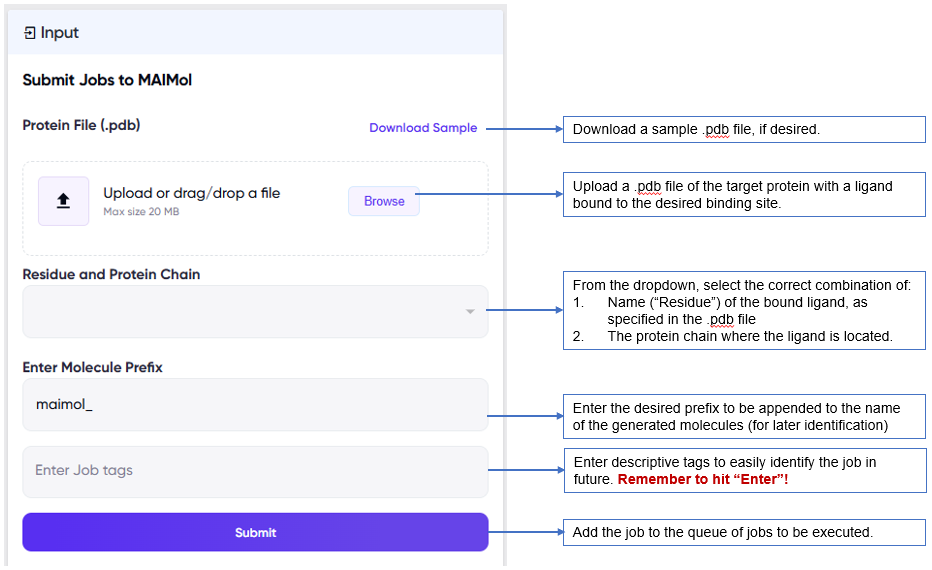
- In the .pdb file supplied, it is necessary to have a ligand bound at the target binding site, since once the user specifies the name of the ligand (“Residue”) and protein chain, MAIMol uses this information to define the binding pocket, and generate molecules accordingly.
- MAIMol currently works correctly only for binding sites made up of residues from a single protein chain only. We will shortly add the capability to handle binding pockets consisting of residues from multiple chains/proteins.