User Inputs
-
Protein Structure (.pdb file): Upload the .pdb file containing the protein structure for which ligands are to be generated. Note that a PDB with a ligand bound to the desired binding pocket is required, as MAIMol 2.0 uses the ligand location for inferring the target site. Select the residue name for the ligand bound to the desired binding site, and the relevant protein chain in the “Residue and Protein Chain” option.
-
Key interactions: Specify the interaction(s) that the generated molecules should have at the binding site. This information will be used to generate molecules with biologically relevant interactions.
The information above is entered through the “Submit Jobs to MAIMol 2.0” fields (Figure 1).
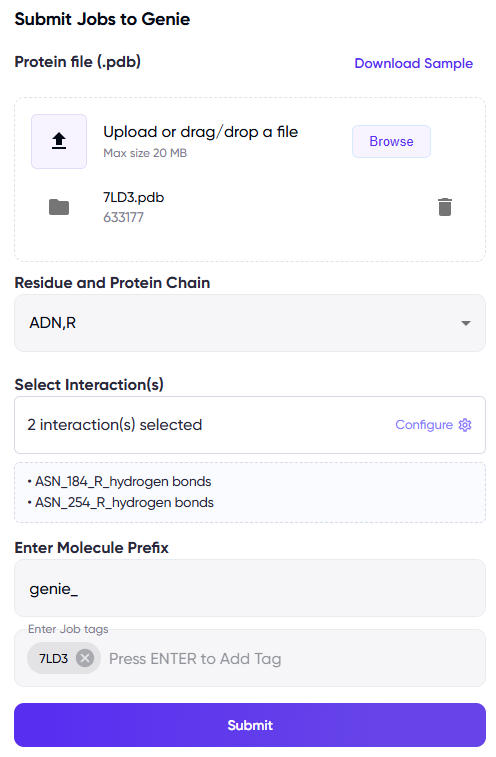
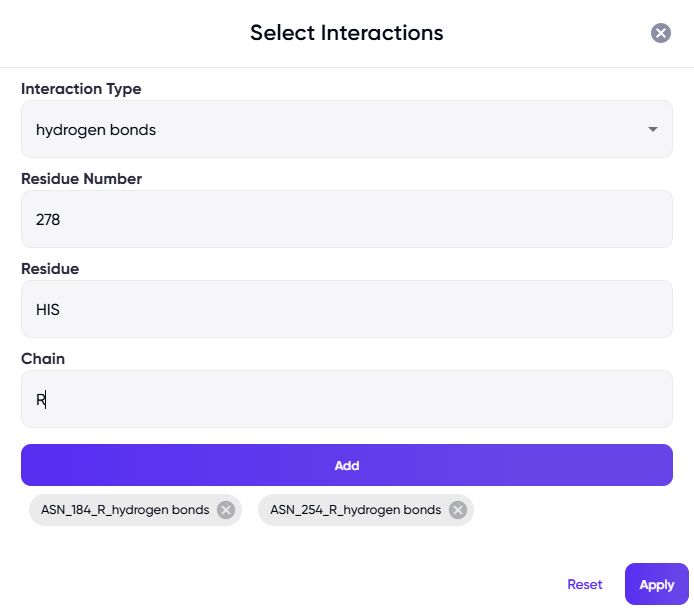
Figure 1 Left panel: Fields for uploading the protein PDB file, specifying the ligand and chain names, and for specifying key interactions. Optionally, prefixes can be specified for adding to the names of the generated molecules, tags can be used to easily locate the job in the jobs list in the future, and a sample PDB file can be downloaded via the “Download Sample” link at the top right of the panel. Right panel: The dialog box that appears upon clicking on the “Select Interaction(s)” field.