User Inputs
The user can input a single SMILES string of the molecule of interest, or a .csv file containing the SMILES string(s) and select the following parameters (Figure 1):
- Properties to optimize: The model can optimize different combinations of Quantitative Estimate of Drug-likeness (QED), Synthetic Accessibility (SA), Molecular Weight (MW), and logP. These are available via the “Choose Properties” dropdown menu.
- Number of molecules to be generated: This is to be entered into the “Enter Number of Molecules…” field, and will be approximately equal to the total number of generated molecules for all input molecules combined.
Note: The number of molecules generated for every input molecule will be approximately equal to the total number generated, divided by the number of input molecules. - “Enter Job Tags” is an optional field to enter descriptive identifiers for convenient future retrieval.
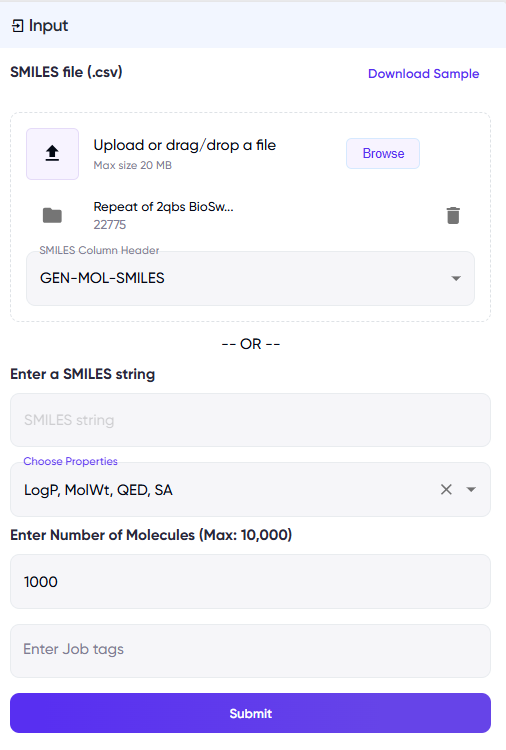
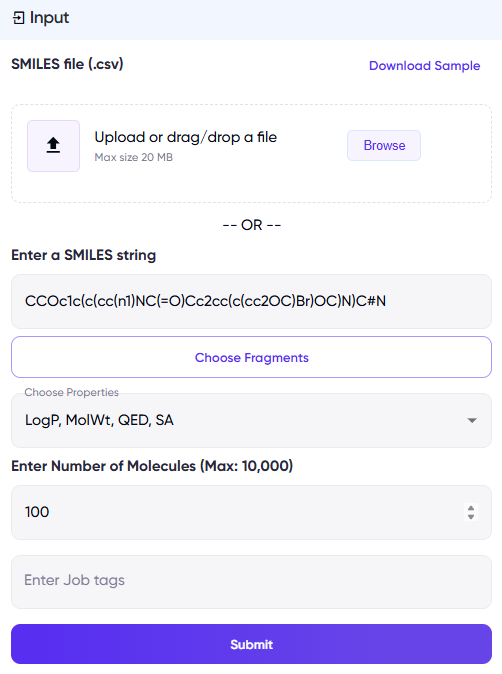
Figure 1: Input fields for BioSwap. The left panel illustrates a screen for the .csv input option, while the right panel illustrates a screen for a single SMILES input.
- For the .csv input option, the user needs to specify the column in the .csv file containing the SMILES strings of interest in the “SMILES column header” field, which appears upon uploading a .csv file (Figure 1, left panel).
- For the single SMILES input option, a “Choose Fragments” button appears upon entering a SMILES string (Figure 1, right panel). Upon clicking this, a list of replaceable fragments appear in a pop-up (Figure 2), and the user can select the fragments they want to replace. If the “Choose Fragments” step is skipped, then molecules will be generated by replacing all possible fragments.
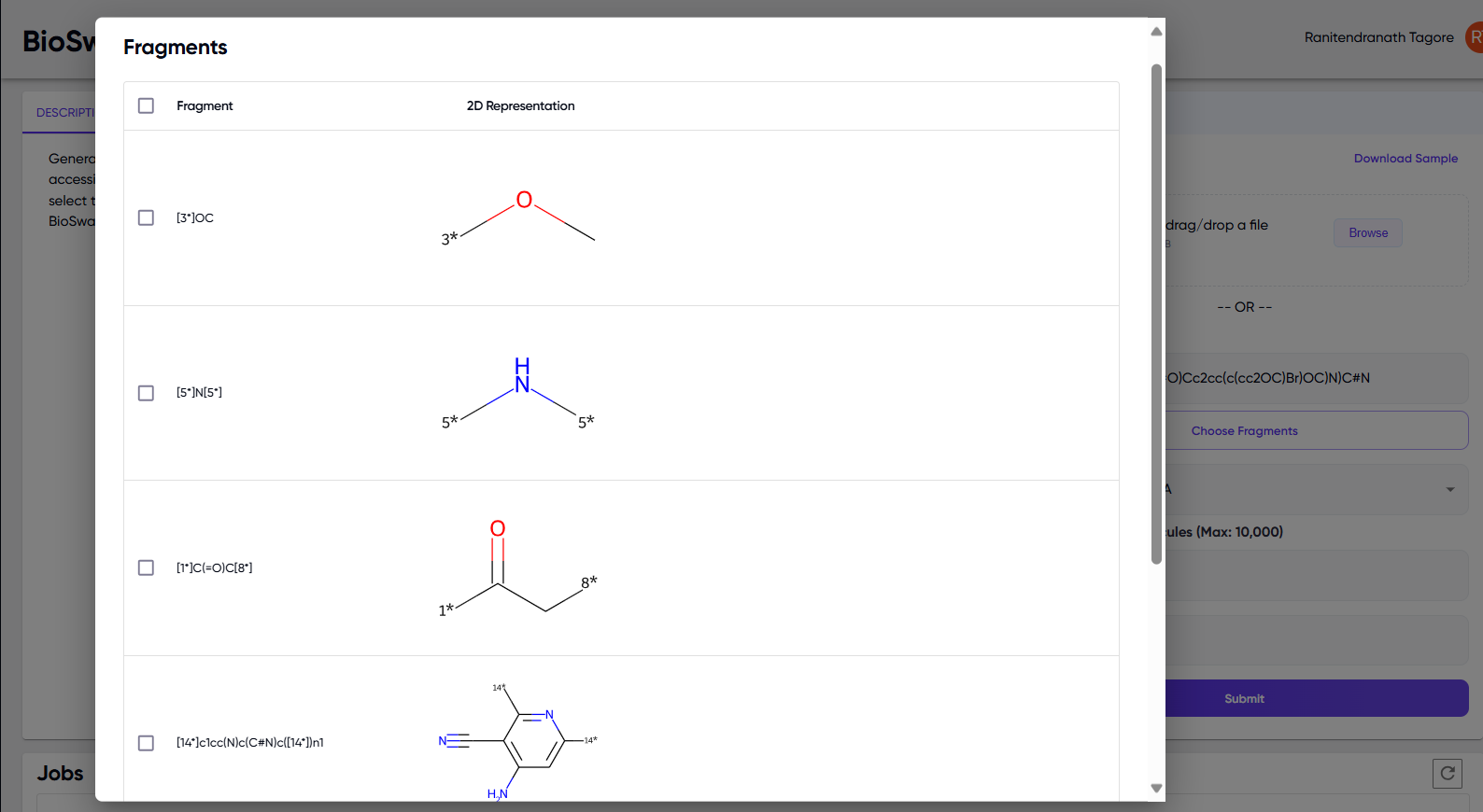
Figure 2: Pop-up of replaceable fragments that appears upon clicking the “Choose Fragments” button for a single SMILES input.